This challenge is over, but if you can beat the top score submit your solution to be featured on the leaderboard.
 Max Quasi-Cliques Challenge
Max Quasi-Cliques Challenge

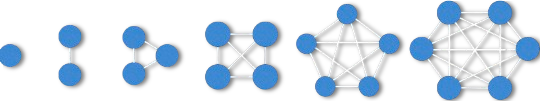
In this data challenge we're looking for dense subgraphs in the FlyWire Brain Connectome (neuronal wiring diagram). Specifically, we consider the simplified undirected and unweighted graph underlying the connectome, where vertices represent neurons and edges represent synaptic connections, and look for dense subgraphs in it. A subgraph with k nodes and m edges is a quasi-clique with density δ (or δ-quasi-clique in short) if at least a δ-fraction of its vertex pairs are connected, that is, if:
m ≥ δ · [k · (k − 1) / 2]
Input & Objective
You’ll download an unweighted, undirected graph G = (V, E). Your goal: extract up to 12 large quasi-cliques, one for each density level:
δi = 1/2i, i = 0,1,...,11
Scoring
- Every δi-quasi-clique is also δi+1-quasi-clique. Hence you can submit fewer than 12 subgraphs and we’ll extend to the implied ones.
- Your solutions for
δ0...δ11yield subgraph sizesk0...k11, and your score isk0/20 + k1/21 + ... + k11/211(higher weights for denser subgraphs). - In case of score ties, earlier submissions rank higher.
Prerequisites
This challenge is open to all. No prior knowledge of neuroscience is needed - just strong data processing and algorithm skills.
Data
- Download the simplified FlyWire connectome graph here; the file contains sorted node pairs (edges); if (x, y) is in the list, (y, x) is also an edge
- Review the sample benchmark solution: here
- Refer to Python code for calculating densities and scoring in this Colab notebook
Submissions
- Make a single CSV file with one line per quasi-clique (up to 12) - similar to the benchmark example
- Each row = comma-separated neuron IDs (one subgraph per row); no headers or other info - our verifier will automatically determine which density each subgraph satisfies
- Email your CSV to arie@princeton.edu
- Please run the verifier before submitting to confirm your score and
kivalues - you may include them in the email for verification - Kindly introduce yourself in your first email, tell us how you learned about the challenge, and whether we can list your name on the leaderboard
- There's no limit on number of submissions — we’ll keep your best one out of those submitted before the deadline
- Winners receive a special trophy from the FlyWire team at Princeton University, and an optional invitation to present (see past winner presentations: 1, 2, 3)
Additional Notes
- Leaderboard shows placements only; all scores and solutions will be published after the deadline
- Challenge updated on July 21, 2025, to allow a range of density values (vs. fixed ones)
- Deadline extended from July 31 to August 31. No further changes or extensions are expected.
This challenge is over, but if you can beat the top score submit your solution to be featured on the leaderboard.
Placement is based on score. Ties (if any) are broken by submission date and marked accordingly.